Assay Assembly for Miniaturized Quantitative PCR in a 384-well Format Using the Echo Liquid Handler
Celeste Glazer, Sammy DatwaniAbstract
Quantitative PCR (qPCR) has enabled a wide range of real-time applications including comparisons in gene expression, genotyping and SNP analysis. To ensure high data quality, the liquid handling employed in low-volume reactions must be exceptionally precise and accurate. This application note highlights the capabilities of the Echo 555 Liquid Handler to dispense into 384-well qPCR plates with total reaction volumes as low as 250 nanoliters. Standard deviations of less than 0.25 and CVs of less than 2% are seen routinely across plates using the low volume dispensing of the Echo Liquid Handler. The ability of the Echo Liquid Handler to transfer from any well to any well simplifies assay setup. The results demonstrate the capabilities to enable assay setup with flexible plate layouts, reduce reagent volumes and increase throughput.
Experiment 1
Quantitative PCR miniaturization in 384-well format
To evaluate the ability to miniaturize qPCR experiments, 250 to 2000 nanoliters of a qPCR reaction mix was transferred from a 384-well Echo Qualified Source Plate into a 384-well qPCR plate (Roche Applied Science). Resulting amplification values were compared.
Methods
Pre-mixed PCR solution containing 1 ng/μL of cDNA (Clontech), 600 nM GAPDH primers (UPL, Roche Applied Science), 400 nM GAPDH probe (UPL, Roche Applied Science), and 1X master mix (Real Time Ready Probes Master, Roche Applied Science) was dispensed into 12 wells of a 384-well Echo qualified polypropylene source plate. The Echo 555 Liquid Handler was then used to transfer 250, 500, and 1000, and 2000 nL of the reaction mix to each quadrant of a 384-well qPCR plate (Roche Applied Science). Reactions were thermal cycled in a LightCycler 480 System (Roche Applied Science). Thermal cycle conditions were as follows: 95°C for 60 seconds, followed by 45 cycles of 95°C for 30 seconds and 60°C for 60 seconds. Amplification results were quantified using the LightCycler 480 system software. Each quadrant of the plate represents a different amount and thus different end-point signal output.
Results
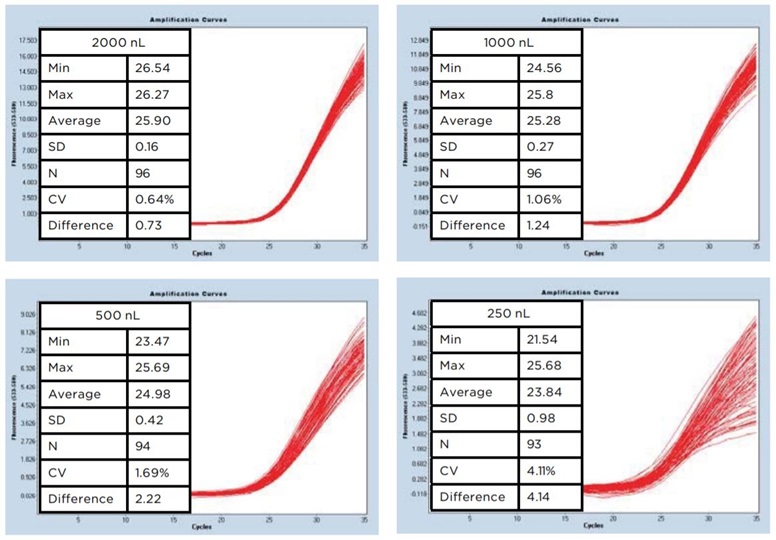
Resulting data demonstrated an increasing endpoint signal which directly corresponded to the total reaction volume in the plate (Figure 1). Excellent standard deviations for 2000 nL and 1000 nL volumes were 0.16 and 0.27, respectively, with coefficients of variation (CV) of 0.64% and 1.06%, respectively. Decreased volume below 1000 nL resulted in higher standard deviation and CVs, with a maximum of 4.11% for a 250 nL reaction volume.
Figure 1: Cp amplification curves at decreasing volumes
Experiment 2
cDNA dilution series using the Echo Liquid Handler for 384-well qPCR
To evaluate the ability of the Echo Liquid Handler to directly dilute cDNA without pre-mixed dilution steps, increasing volumes of concentrated stock cDNA were transferred into a 384-well qPCR destination plate to create a series dilution.
Methods
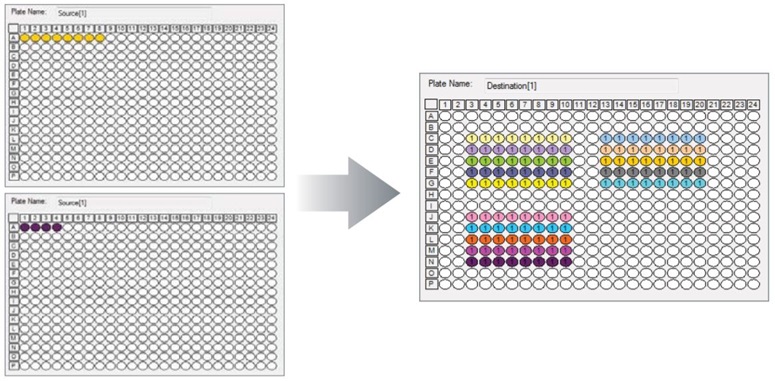
Using a two-step process, the Echo Liquid Handler was used to dispense master mix with premixed primers and probes into a qPCR plate, followed by incremental transfer of stock human reference cDNA. Echo Plate Reformat software was used to set up the transfer of master mix and cDNA (Figure 2). Source plates containing 50 μL of premixed master mix, primers and probes (600 nM and 400 nM GAPDH respectively) were pipetted into eight wells of an Echo qualified 384-well polypropylene source plate. A protocol was created using Echo Plate Reformat to dispense incremental fill volumes of master mix (see Table 1). A second source plate (low dead volume, Echo qualified) containing human reference cDNA at a concentration of 50 ng/μL was used to create the dilution series. Echo Plate Reformat software was set up to transfer a four-fold dilution series from 2.5 nL to 640 nL. After the protocols were created, the actual dispense time for both plates totaled less than 8 minutes.
| Final cDNA Concentration | 0.13 ng | 0.5 ng | 2.0 ng | 8.0 ng | 32.0 ng |
| Master mix (nL) | 2000 | 2000 | 2000 | 1840 | 1360 |
| cDNA (nL) | 2.5 | 10 | 40 | 160 | 640 |
| Total | 2002.5 | 2010 | 2040 | 2000 | 2000 |
Table 1: Dispense volumes for cDNA dilution series qPCR
Figure 2: Echo Plate Reformat software plate maps for cDNA dilution series qPCR. Top left: Master mix containing primers and probes at 1X concentration. Bottom left: Stock cDNA at 50 ng/μL. Right: Reformatted plate.
Results
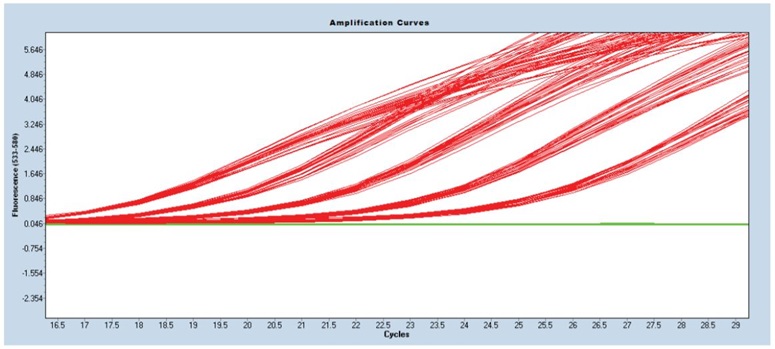
Resulting amplification curves demonstrated a distinct delineation between concentration and cycle amplification (Cp) value for each change in concentration (Figure 3 and Table 2). Data analysis for each of the 24 replicates showed excellent standard deviations from 0.085-0.18 with CVs of the Cp ranging from 0.50-0.73%.
Figure 3: Cp amplification curves
| cDNA Amount | 0.13 ng (2.5 nL) | 0.5 ng (10 nL) | 2.0 ng (40 nL) | 8.0ng (160 nL) | 32.0 ng (640 nL) |
| Cq Min | 24.66 | 22.84 | 20.83 | 19.11 | 17.05 |
| Cq Max | 25.63 | 23.34 | 21.48 | 19.63 | 17.33 |
| Cq Average | 25.11 | 23.09 | 21.15 | 19.31 | 17.17 |
| SD | 0.18 | 0.13 | 0.17 | 0.15 | 0.09 |
| N | 24 | 24 | 24 | 24 | 24 |
| CV | 0.73% | 0.55% | 0.80% | 0.80% | 0.50% |
| Difference | 0.97 | 0.5 | 0.65 | 0.52 | 0.28 |
Table 2: Data analysis for a cDNA dilution series
Experiment 3
qPCR assay assembly using Echo Plate Reformat software
The Echo Liquid Handler was used to generate a complete PCR from individual components.
Methods
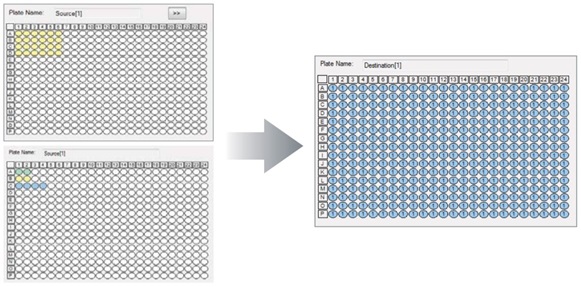
Two Echo Qualified polypropylene source plates were used; one plate (384PP_AQ_SP) to dispense the master mix, and a second low dead volume plate (384LDV_AQ_B) to dispense primers, probes, and cDNA. First, 1500 nL of a 1X master mix was transferred to the 384-well qPCR plate. This was followed by 40 nL of primers (20 μM) and 40 nL of probe (10 μM). Thirty nL of alternating cDNA (~50 ng/μL) or water controls were added, as shown in Figure 4. In this experiment, alternating amounts of water and of cDNA were dispensed to mimic zero cross contamination experimental conditions. Concentrations of stock solution were 20 μM GAPDH primers, GAPDH 10 μM, and 50 ng/mL of human reference cDNA. Final concentrations were 1.5 ng cDNA, 0.53 μM primers, 0.267 μM probe.
Figure 4: Plate Reformat software setup for multi-step qPCR reagent transfer. Source plates: Top left, mastermix and bottom left, Row A: Primers, Row B: Probes, Row C: cDNA.
At right, PCR plate with 1500 nL mastermix/well. Half of wells contain primers, probes and cDNA. Remaining wells contain water as a control.
Results
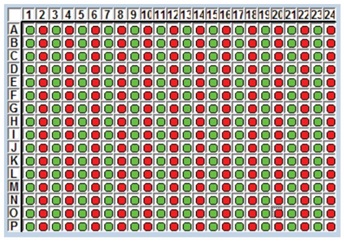
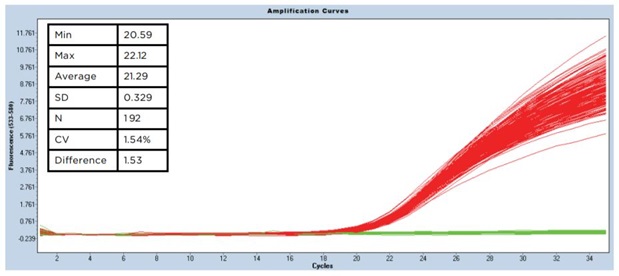
The heat map results displayed in Figure 5 show the positive and negative calls from the software analysis of the Cp values. The positive calls are in shown in green and the negative calls are shown in red. The precise non-contact transfer of the Echo Liquid Handler enabled zero cross contamination despite a four step dispense. Figure 6 shows the amplification results as well as the standard deviation and CVs for 192 positive samples. The standard deviation was 0.32 with a CV of 1.54% for a 1500 nL final volume.
Figure 5: Heat map results from qPCR assay assembly
Figure 6: Amplification results of qPCR assay assembly
Summary
The Echo liquid handling platform presents exciting new capabilities for total assembly and miniaturization of qPCR in 384-well formats. The ability of the Echo Liquid Handler to transfer from any well to any well enables experiment assembly utilizing concentrated stock material without the typical pre-pipetting steps required to dilute source solutions. Superior volumetric precision and accuracy ensure excellent cycle quantification precision even with very little target DNA in very low reaction volumes. This technology enables scientists to fully explore the capabilities of miniaturized PCR and other genomics applications, while reducing reagent and consumable costs.